DeepTCR: A Deep Learning Framework for Revealing Structural Concepts within TCR Repertoire

Abstract
Background: Artificial intelligence is poised to revolutionize every aspect of human life, finding applications in everything from self-driving cars to diagnosing cancer. In fact, almost any task that involves pattern recognition can be formulated in a way that modern AI algorithms can be used to achieve super-human performance. The immune synapse is a highly complex interaction between several proteins and peptides that allows for a constant surveillance of foreign invaders. However, modeling these interactions is extremely difficult as the combinations of interactions is simply intractable. In immuno-oncology, the study of this interaction is crucial as anti-tumor responses rely on sensitive and specific recognition of tumor-specific antigens. Implications of accurately predicting and modeling these interactions in immune-oncology range from improved and potent vaccine design to biomarkers for predicting response to immunotherapy. Methods: Our group has developed a variety of deep learning models to model the signal transmission within the immune synapse. At the core of all architectures designed, convolutional layers, similar to ones used to learn features in images, are used to learn motifs within the sequencing data for a predictive or descriptive purpose. Results: We first present AI-MHC, an applied deep convolutional neural network for class-specific MHC binding algorithm that achieves state-of-the-art performance in both Class and Class II predictions (Figure 1A,B)1. By incorporating ‘meaning’ of the allele within the network, we are able to model the interaction of allele and peptide within the context of a neural network (Figure 1C). We take these concepts further in the development of DeepMANA, a deep learning framework which combines sequence-specific information about an allele/peptide pairing to not only predict binding affinity for any allele with a known protein sequence but also provide an antigen ‘quality’ score, based on the “non-self/foreign-ness” of a neoantigen. We observe in three previously published immunotherapy clinical trials, these quality neoantigens are enriched in long-term survivors or responders (Figure 2)2. Finally, we present DeepTCR, a collection of unsupervised and supervised deep learning algorithms capable of revealing structure in T-cell receptor repertoire. We demonstrate that DeepTCR achieves state-of-the-art performance in clustering antigen-specific TCR’s (Figure 3A) and is capable of learning a predictive signature in TIL repertoire of mice treated with various immunotherapies (Figure 3B)3,4. Conclusion: These types of AI technologies could yield an entire new area of biomarker discovery as well as improve our understanding of the complex interaction occurring at the immune synapse that is ultimately required for a successful anti-tumor response.
Figure 1
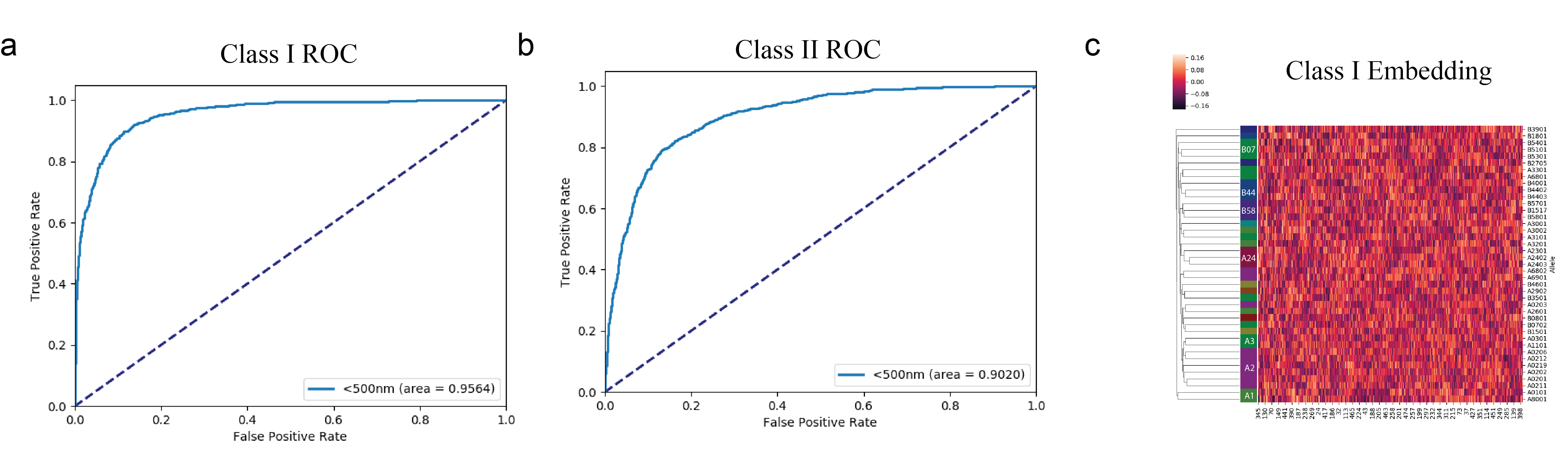 AI-MHC: an allele-integrated deep learning framework for improving Class I & Class HLA-binding predictions. a-b) Receiver Operator Characteristic curves for predicting Class I & Class II binding peptides. c) Trainable embedding layer for HLA alleles was extracted from neural network to examine the vectorization of each allele. Our results reveal that the network is able to cluster HLA alleles of the same supertype, suggesting it has learned from the data which alleles share similar peptide binding properties.
AI-MHC: an allele-integrated deep learning framework for improving Class I & Class HLA-binding predictions. a-b) Receiver Operator Characteristic curves for predicting Class I & Class II binding peptides. c) Trainable embedding layer for HLA alleles was extracted from neural network to examine the vectorization of each allele. Our results reveal that the network is able to cluster HLA alleles of the same supertype, suggesting it has learned from the data which alleles share similar peptide binding properties.
Figure 2
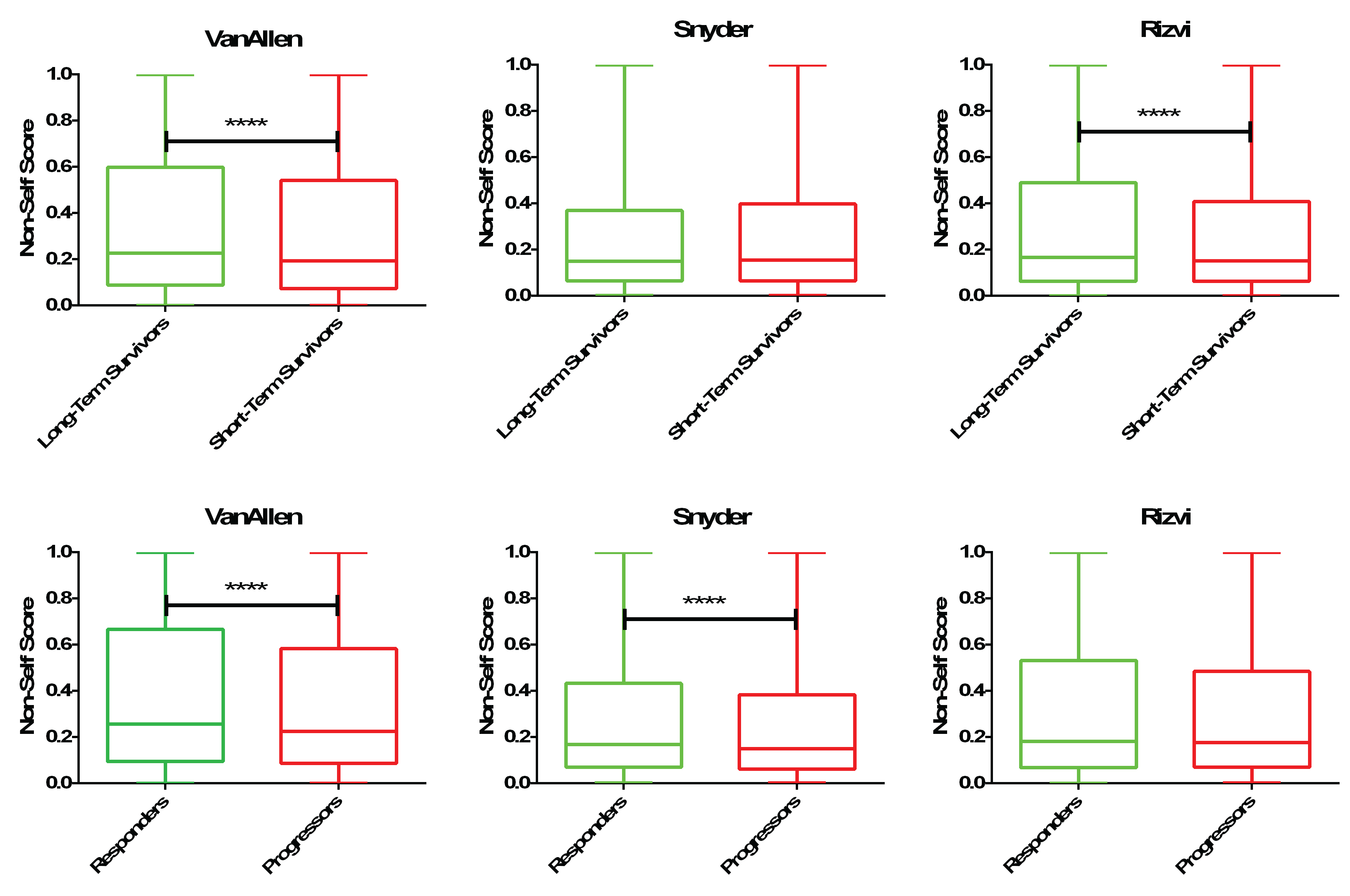 DeepMANA: a deep learning framework for prediction of ‘high-quality’ neoantigens. DeepMANA was used to predict the quality scores of all neoantigens for long/short term survivors and responder/progressors to immunotherapy. We note enrichment in either long-term survivors or responders for ‘high-quality’ neoantigens, those who are characterized by the neural network to be non-self/foreign in nature.
DeepMANA: a deep learning framework for prediction of ‘high-quality’ neoantigens. DeepMANA was used to predict the quality scores of all neoantigens for long/short term survivors and responder/progressors to immunotherapy. We note enrichment in either long-term survivors or responders for ‘high-quality’ neoantigens, those who are characterized by the neural network to be non-self/foreign in nature.
Figure 3
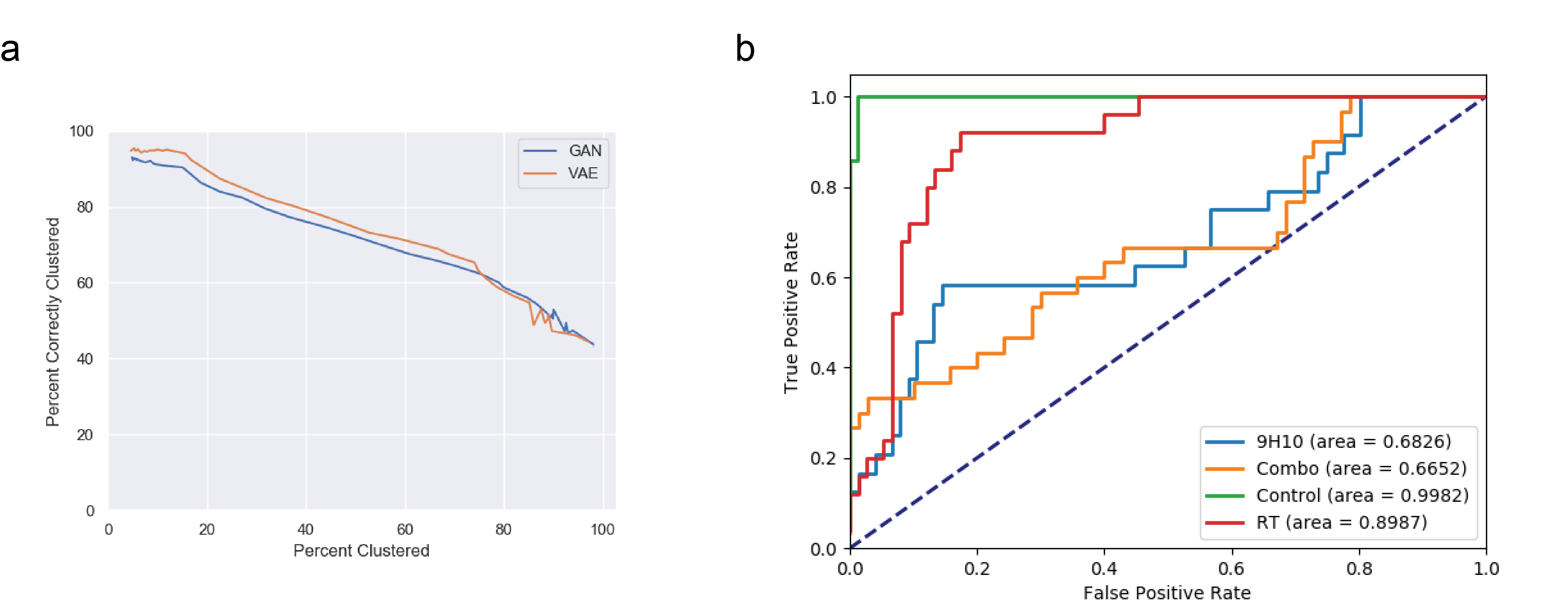 DeepTCR: a deep learning framework for revealing structural concepts in TCR repertoire. a) A previously published dataset of 2067 sequences spanning 7 specificities from tetramer+ sorted cells were collected and two unsupervised deep learning approaches (VAE & GAN) were used to cluster these sequences. Our algorithm demonstrates state-of-the-art ability to specifically cluster antigen-specific cells while maintaining a high sensitivity. b) TCRSeq data from a previously published dataset was collected where four cohorts of tumor-bearing mice were harvested for their tumor-infiltrating lymphocytes. DeepTCR learned a strong predictive signature of the mice that received no treatment and those who had received radiation therapy alone.
DeepTCR: a deep learning framework for revealing structural concepts in TCR repertoire. a) A previously published dataset of 2067 sequences spanning 7 specificities from tetramer+ sorted cells were collected and two unsupervised deep learning approaches (VAE & GAN) were used to cluster these sequences. Our algorithm demonstrates state-of-the-art ability to specifically cluster antigen-specific cells while maintaining a high sensitivity. b) TCRSeq data from a previously published dataset was collected where four cohorts of tumor-bearing mice were harvested for their tumor-infiltrating lymphocytes. DeepTCR learned a strong predictive signature of the mice that received no treatment and those who had received radiation therapy alone.