Abstract
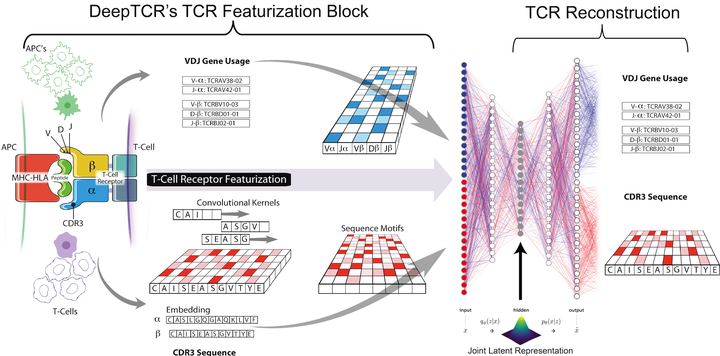
Deep learning algorithms have been utilized to achieve excellent performance in pattern-recognition tasks, such as in image and vocal recognition$^textrm1,2$. The ability to learn complex patterns in data has tremendous implications in the genomics world, where sequence motifs become learned ‘features’ that can be used to predict functionality, guiding our understanding of disease and basic biology$^textrm3–6$. T-cell receptor (TCR) sequencing assesses the diversity of the adaptive immune system, and while prior conventional biological sequence analysis tools have been insightful, they have significant shortcomings. Prior approaches have been limited to single-sequence analytics and thus unable to characterize the overall structural information within an entire sample of sequences. Furthermore, they utilize only unsupervised approaches, being unable to leverage labels to guide the learning process$^textrm7–9$. We present DeepTCR, a broad collection of unsupervised and supervised deep learning methods able to uncover structure in highly complex and large TCR sequencing data. We demonstrate its utility across multiple scientific examples, including learning antigen-specific motifs to viral and tumor-specific epitopes and understanding immunotherapy-related shaping of repertoire. We further extract meaningful motifs from the trained network as a means of explaining the sequence concepts that have been learned to accomplish a given task. Finally, we extend DeepTCR’s functionality to analyze paired α/β chains as inputs, demonstrating the ability to query the contribution of each chain to antigen-specificity. Our results show the flexibility and capacity for deep neural networks to handle the complexity of high-dimensional genomics data for both descriptive and predictive purposes.