AI-MHC: an allele-integrated deep learning framework for improving Class I & Class II HLA-binding predictions
Abstract
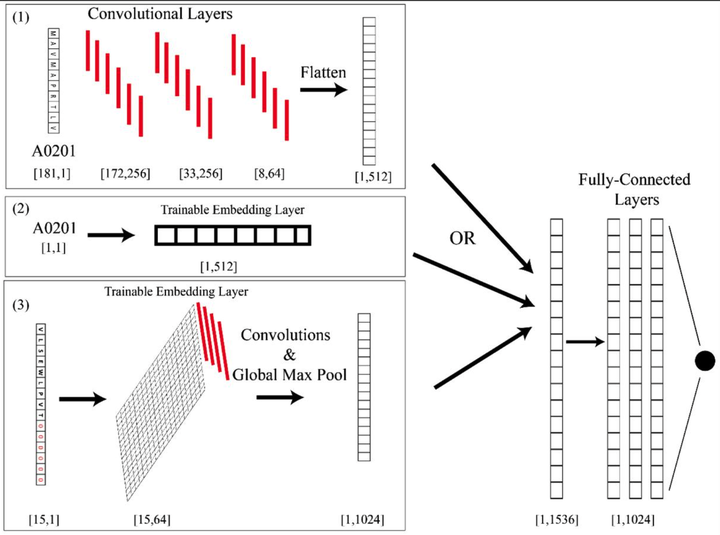
The immune system has potential to present a wide variety of peptides to itself as a means of surveillance for pathogenic invaders. This means of surveillances allows the immune system to detect peptides derives from bacterial, viral, and even oncologic sources. However, given the breadth of the epitope repertoire, in order to study immune responses to these epitopes, investigators have relied on textitin-silico prediction algorithms to help narrow down the list of candidate epitopes, and current methods still have much in the way of improvement.textless/ptextgreatertextlessh3textgreaterResultstextless/h3textgreater textlessptextgreaterWe present Allele-Integrated MHC (AI-MHC), a deep learning architecture with improved performance over the current state-of-the-art algorithms in human Class I and Class II MHC binding prediction. Our architecture utilizes a convolutional neural network that improves prediction accuracy by 1) allowing one neural network to be trained on all peptides for all alleles of a given class of MHC molecules by making the allele an input to the net and 2) introducing a global max pooling operation with an optimized kernel size that allows the architecture to achieve translational invariance in MHC-peptide binding analysis, making it suitable for sequence analytics where a frame of interest needs to be learned in a longer, variable length sequence. We assess AI-MHC against internal independent test sets and compare against all algorithms in the IEDB automated server benchmarks, demonstrating our algorithm achieves state-of-the-art for both Class I and Class II prediction.textless/ptextgreatertextlessh3textgreaterAvailability and Implementationtextless/h3textgreater textlessptextgreaterAI-MHC can be used via web interface at baras.pathology.jhu.edu/AI-MHC