
Introduction
This story begins at the Annual Association for Cancer Research (AACR) 2016 Annual Meeting. I was attending a talk being given by Google showing the advances they had made in deep learning and the potential applications within the medical space. I was so captivated by the power of deep learning at the time (and saw where I could apply it within my work in TCR repertoire analyis - more on this later), that before the talk was over, I had purchased THE textbook on deep learning and would go through it along with earning a Udacity Nanodegree in deep learning.
ICIAR 2018 Challenge
After I had gone through the textbook on deep learning and the Udacity Nanodegree, I thought in order to really put these new skills to the test, I should participate in one of the machine learning challenges. With that, me and my co-mentor Alexander Baras entered into the ICIAR 2018 Challenge which was focused around diagnosing breast cancer histology. With Alex’s background as a pathologist, I was able to learn about the nuances and practical aspects of using deep learning in medical images.
Model
Despite the promise of deep learning for applications in the medical field, due to the novelty of these applications, there exists little labeled data for supervised machine learning. However, prior work in the fields of deep learning has shown the power of a technique called ‘transfer learning,’ the idea being that pre-existing designed architectures (i.e. ResNet50, AlexNet, VGG16) that have been trained on possibly millions of images for over 1000 image classes can serve as ‘professional feature detectors’ for new visual recognition tasks where data may not be as abundant. While it may seem far fetched that a convolutional neural network (CNN) trained to recognize dogs and cats could recognize relevant features in medical imaging, Sebastian Thrun and his group demonstrated this exact concept could be utilized, generating results even better than when training a CNN de-novo to diagnose skin lesions. In our manuscript, we propose a method by which one can use ResNet50, a pre-trained CNN, as a feature detector for classification of normal, benign, in-situ, and invasive breast carcinoma. We propose a method by which convolving this pre-trained net at various magnifications of labeled pathology image tiles can serve to detect low and high-level features in digital pathology that can be ultimately used for the task of lesion classification.
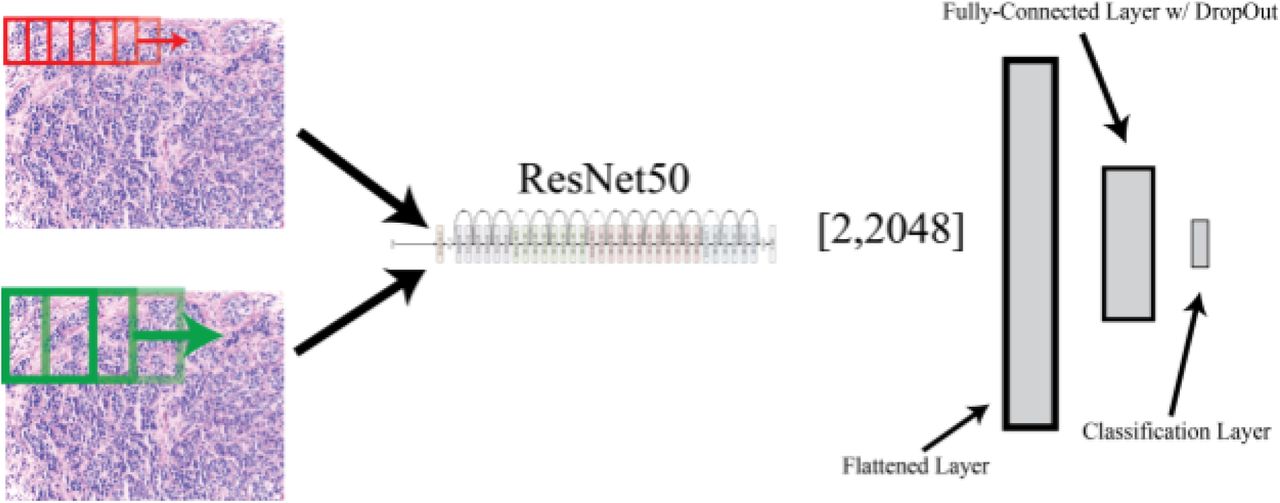
Results
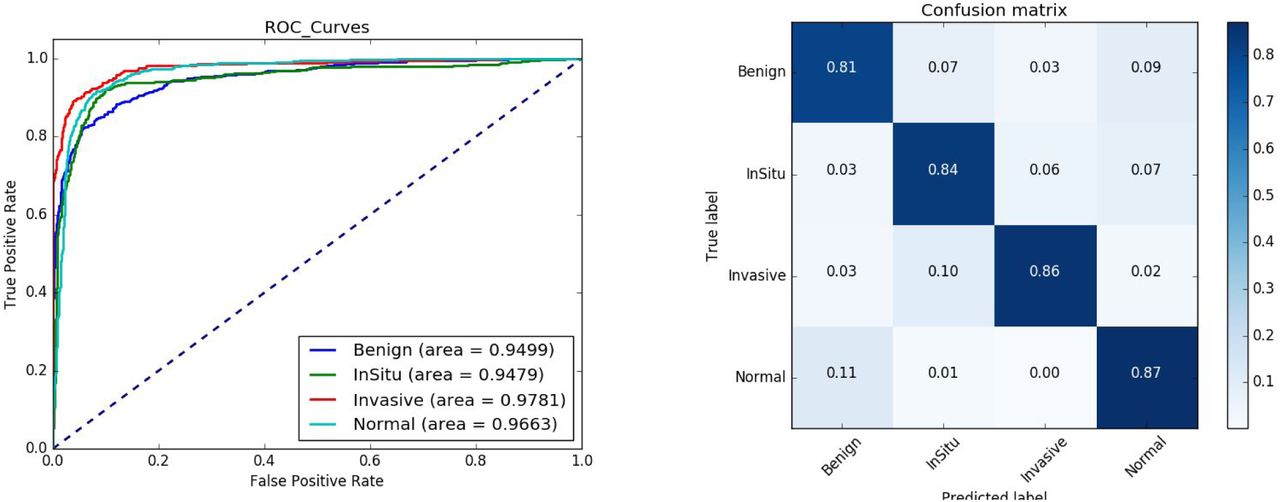
In our manuscript, we explore multiple methods to improve performance and generalizability of our model including image augmentation. In essence, we demonstrate our model achieves a high level of performance in differentiating between these different forms of breast histology. In the challenge, our model came in 16th place in the world and 3rd place in the United States.
Reflection
While this project was not formally part of my doctoral work, it was my first hands-on experience and exposure to deep learning. This became the first step in using deep learning towards my study of T-cell repertoires, which became the original reason I wanted to learn how to implement methods in deep learning.